ICGC-TCGA DREAM Somatic Mutation Calling Challenge
4.9 (207) · $ 19.50 · In stock
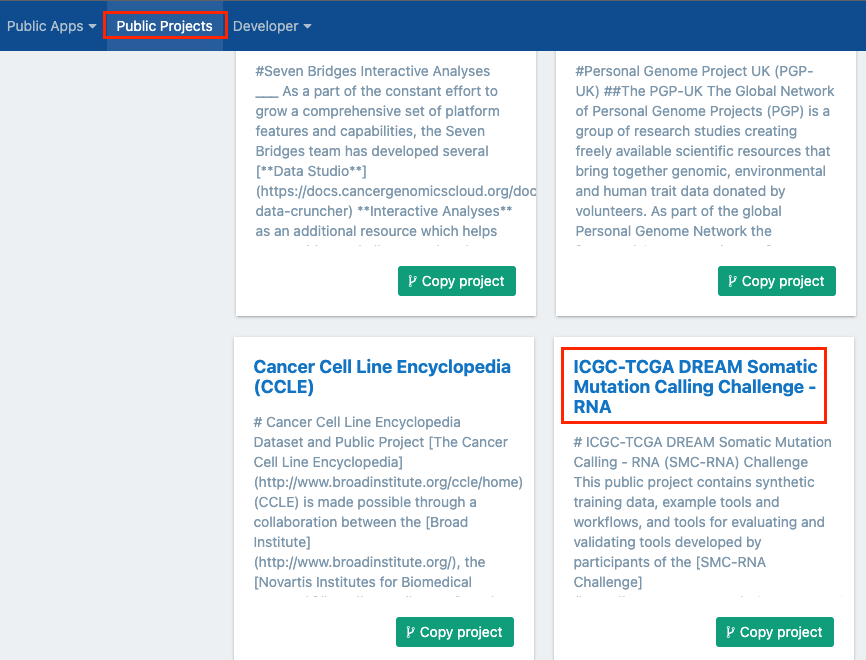
🚧On this page:OverviewAccess ICGC-TCGA DREAM Somatic Mutation Calling Challenge resources on the CGCCopy the ICGC-TCGA DREAM Somatic Mutation Calling public projectLearn moreWebinar: Visual interfaceWebinar: Python and APIResources OverviewThe Seven Bridges CGC is proud to launch the ICGC-TCGA DREA
Prioritisation of structural variant calls in cancer genomes [PeerJ]
Validating multiple cancer variant callers and prioritization in tumor-only samples

Indel performance of Lancet and other methods on the synthetic tumor #4

Free Bio-IT World Webinar: Machine Learning to Detect Cancer Variants

First challenge to make use of the new NCI Cloud Pilots – Somatic Mutation Challenge – RNA: Best algorithms for detecting all of the abnormal RNA molecules in a cancer cell
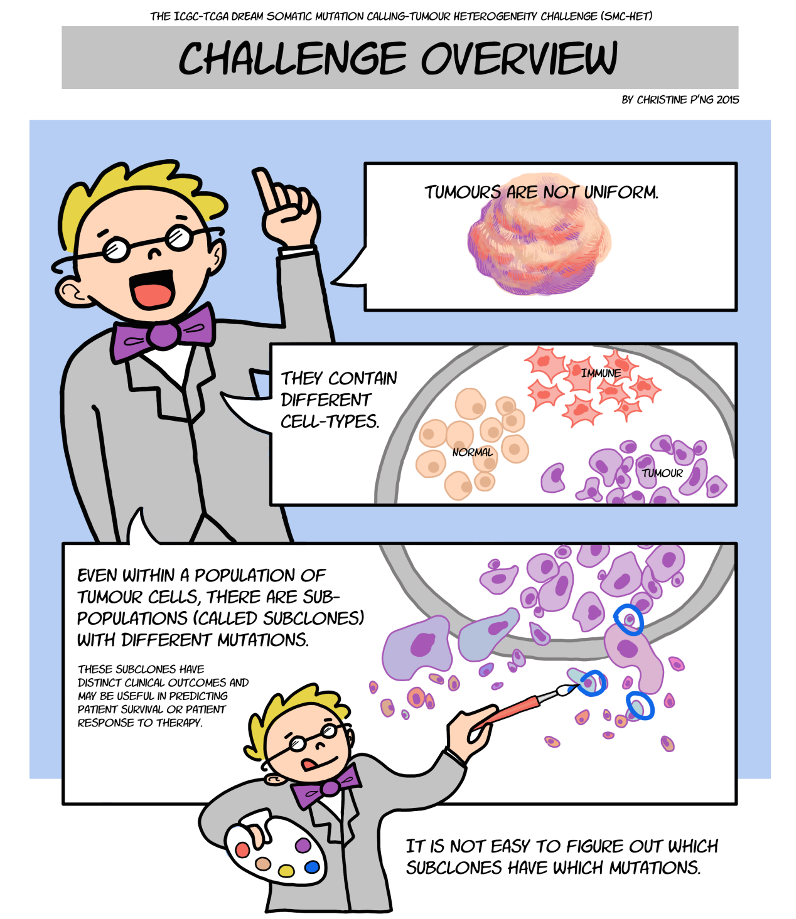
Crowdsourcing Cancer Challenge Donnelly Centre for Cellular and Biomolecular Research

Crowd-sourced benchmarking of single-sample tumour subclonal
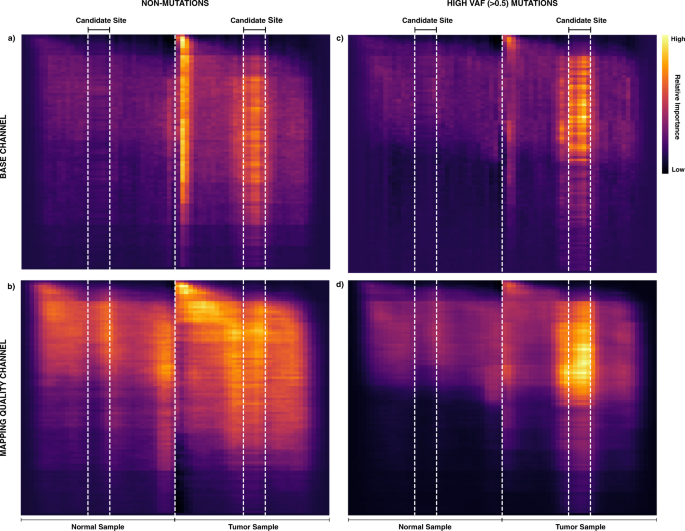
Accurate somatic variant detection using weakly supervised deep

SENTIEON®, DFETECH

Current perspectives on mass spectrometry-based immunopeptidomics: the computational angle to tumor antigen discovery

Improving somatic variant identification through integration of

DREAMTools: a Python package for scoring

PDF) Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data

1 Cancer Sequencing Quality & The ICGC-TCGA DREAM Somatic Mutation

Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines - ScienceDirect